Saquinavir
A peptidomimetic HIV protease inhibitor.
General information
Saquinavir is a human immunodeficiency virus (HIV) protease inhibitor. It is used for treatment and prevention of HIV infection and the acquired immunodeficiency syndrome (AIDS) (LiverTox).
Saquinavir on DrugBank
Saquinavir on PubChem
Saquinavir on Wikipedia
Synonyms
Invirase
Marketed as
INVIRASE (SAQUINAVIR MESYLATE)
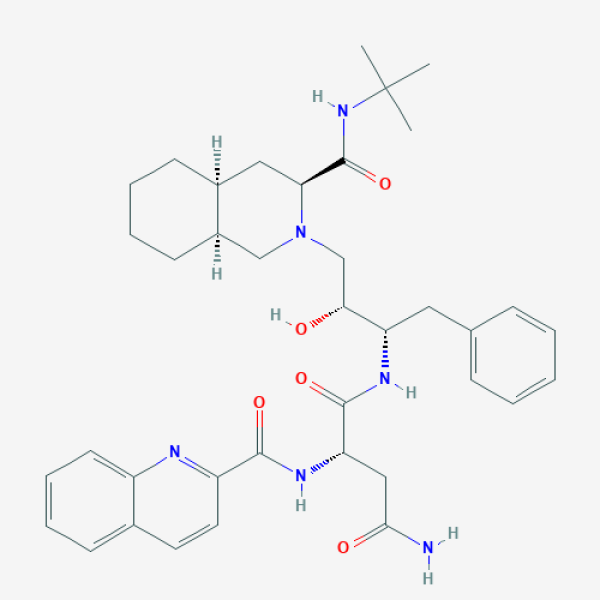
CC(C)(C)NC(=O)[C@@H]1C[C@@H]2CCCC[C@@H]2CN1C[C@H]([C@H](CC3=CC=CC=C3)NC(=O)[C@H](CC(=O)N)NC(=O)C4=NC5=CC=CC=C5C=C4)O
