N3
A tripeptide SARS-CoV-2 3C-like protease inhibitor.
General information
N3 is a SARS-CoV-2 3C-like protease covalent inhibitor with anti-SARS-CoV-2 properties (Jin et al., 2020).
N3 on PubChem
Synonyms
Inhibitor N3; E)-(4S,6S)-8-Methyl-6-((S)-3-methyl-2-{(S)-2-[(5-methyl-isoxazole-3-carbonyl)-amino]-propionylamino}-butyrylamino)-5-oxo-4-((R)-2-oxo-pyrrolidin-3-ylmethyl)-non-2-enoic acid benzyl ester
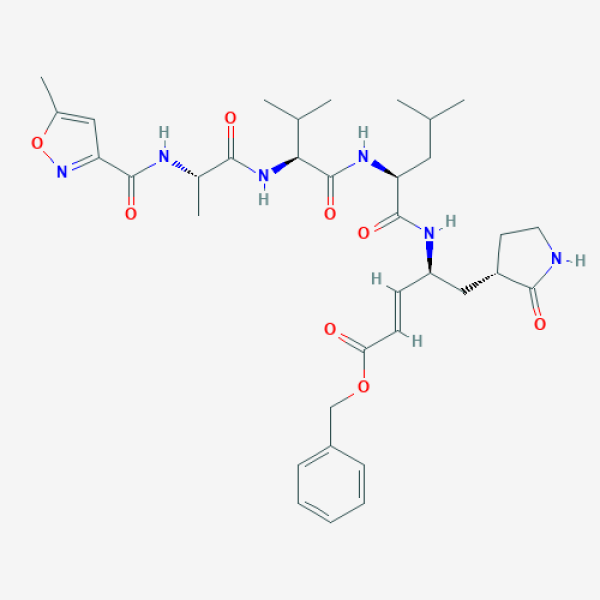
CC1=CC(=NO1)C(=O)N[C@@H](C)C(=O)N[C@@H](C(C)C)C(=O)N[C@@H](CC(C)C)C(=O)N[C@@H](C[C@@H]2CCNC2=O)/C=C/C(=O)OCC3=CC=CC=C3
Supporting references
| Link | Tested on | Impact factor | Notes | Publication date |
|---|---|---|---|---|
|
Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors
3CLpro Crystallization Enzyme assay In vitro In silico |
in silico; in vitro enzyme assay; crystallization; Vero E6 cells | 42.78 | Computationally predicted to form a covalent bond with the SARS-CoV-2 3C-like protease's active site. This bond was experimentally observed after co-crystallization of the compound with the protease. N3 inhibited SARS-CoV-2 infection in Vero E6 cells with EC50 of ca. 16.77 μM. |
Jun/11/2020 |
AI-suggested references
Clinical trials
| ID | Title | Status | Phase | Start date | Completion date |
|---|---|---|---|---|---|
| NCT04924842 | COVID-19 - SARS-CoV-2 Community Contamination in Children and Adults - Impact of Variants (Dyn3CEA - Nosocor Phase 2) | Completed | Jun/22/2021 | Oct/28/2021 | |
|
|||||
| NCT04664296 | COVID-19 - SARS-CoV-2 Community Contamination in Children and Adults (Dyn3CEA_Nosocor) | Completed | Dec/21/2020 | Mar/23/2021 | |
|
|||||
